Structural clarification
New methods could speed up protein research considerably
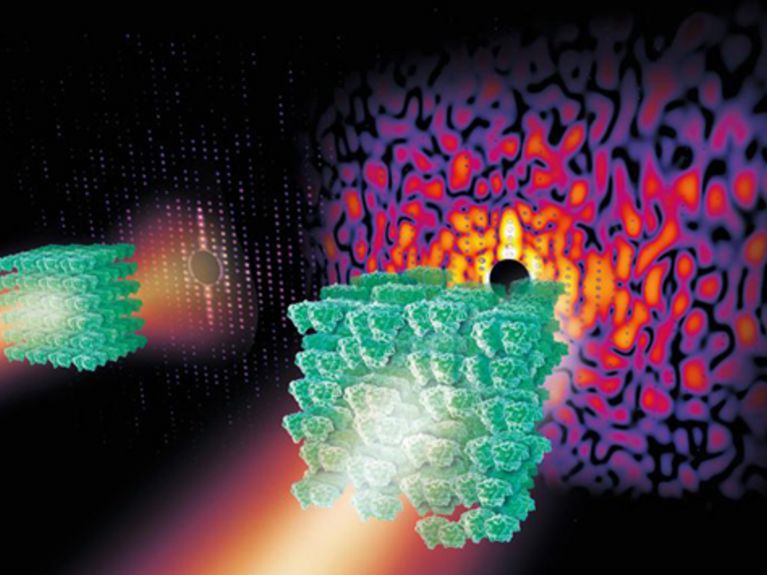
Slightly disordered crystals from complex biomolecules such as the Photosystem II shown here produce a continuous spread pattern in X-ray light from which more information can be obtained than from the so-called Bragg-peaks of a more ordered crystal (left). Image: DESY, Eberhard Reimann
A research team under the leadership of Henry Chapman from the Deutsches Elektronen-Synchrotron (DESY) have developed a new method for clarifying the make-up of biomolecules. It could considerably improve and simplify, in the future, the complicated technology of structural analysis by means of crystallography. The research scientists published their findings in the trade journal Nature.
Herr Chapman, why is a physicist interested in bio-molecules?
We physicists are also interested in these molecular machines that power everything. They are the substance of life. Their long chains made of polymers form fascinating patterns. Particularly, I would like to provide biologists with improved tools of the trade, in order to be able to decipher the spatial structure of these molecules, thus enabling us to better understand the mechanisms of life.
What makes it so difficult to examine biomolecules – why can’t they just be placed under a microscope?
In order to understand molecules, one has to be able to see them very clearly – we would like to know how all of the individual atoms are arranged so that we can understand the molecular structure. This can’t be done with a normal light microscope, since the wavelength is simply too long and the resolution is insufficient. One needs light with a very short wavelength – X-rays. The problem here is that X-rays are highly energised and can kick electrons out of the atoms. This can destroy the objects. Imagine it this way – we’ve got a secret message on a piece of paper and we want to read the message. For this purpose we have to turn the light on. Unfortunately, the message is very sensitive to light and is destroyed in the process.
But how can we get around this problem to read the message?
We do it by means of a detour, in that we grow crystals from the molecules and illuminate them with X-rays. The crystal grid scatters the rays. Through regular arrangement of the molecules in the grid, a characteristic pattern emerges consisting of bright points – the Bragg peaks. It looks quite beautiful. But because we don’t have lenses at an atomic level in order to see patterns, we have to use a computer. It ascertains the structure of the crystal from the position and intensity of the bright points in the X-ray spread pattern. It has thus been possible to determine the structure of tens of thousands of proteins and other biomolecules. The real challenge here is that they have to be crystallised beforehand.
Why is that a problem?
Because not every biomolecule can be easily cultivated as a crystal. Scientists have, to date, invested a lot of blood, sweat and tears into the cultivation of crystals. Some of them have spent more than ten years on the process of growing a crystal from one single protein. And frequently only small, badly arranged crystals emerge, which don’t reveal very much information about the structure of the molecule – at least, this is what research scientists have always thought until now.
Could it be possible to change that with your discovery?
Yes, since we’ve found out that even the disorderly crystals weed out something that reveals the structure of the crystal. It is the weak, continuous spread pattern that we previously weren’t observing. You can imagine it this way – it is like many individual images of a molecule lying on top of one another. It is the equivalent of a disruptive background.
But spread patterns have been investigated for many years now, haven’t they?
Yes, and that is the strange thing about it – because it’s really so obvious. We use the spread patterns to gain insights into the vibrations and movements of the molecules within the crystal, but we haven’t been keeping track of that until now for structural analysis.
Is that comparable to the discovery of junk DNA genetic information?
Yes, it’s just like that. It was there the whole time, but we just never saw it because it was just one pattern among many.
So, it’s a breakthrough?
I hope so. We have now tested our method in a membrane-protein complex called Photosystem II, which is a part of the photosynthesis mechanism in green plants. And now we have the impression that there is even more to it than we thought. It is our goal now to get even higher resolution images and many other macromolecules from the Photosystem II. It could advance and speed up protein research quite a bit.
Could this revolutionise biomolecular research?
At least now we know that we have enough information from the readings. We just have to send the results through a simple algorithm, and it will ascertain the structure. That is the crucial advancement to be able to speed things up. And now we want to find out how comprehensively this method can be implemented. That’s why we are looking at other systems as well – such as the membrane proteins that play such an important role in numerous biological processes and which at least half of all pharmaceuticals are targeted at.
What does this mean for you?
This represents a huge success for me and my team. Now we want to develop these algorithms further and establish the method for structural biologists. Crystallography has been more or less a black box in science for the last ten years; you insert a crystal and maybe you’ll get a structure, or maybe not. I hope that this method will also re-focus on the miracle of structural analysis and expand our view of the fundamental mechanisms of life.
Macromolecular diffractive imaging using imperfect crystals; Kartik Ayyer et al. „Nature”, 2016; DOI: 10.1038/nature16949
Readers comments