SARS-CoV-2
Blueprint for an active substance against the novel coronavirus
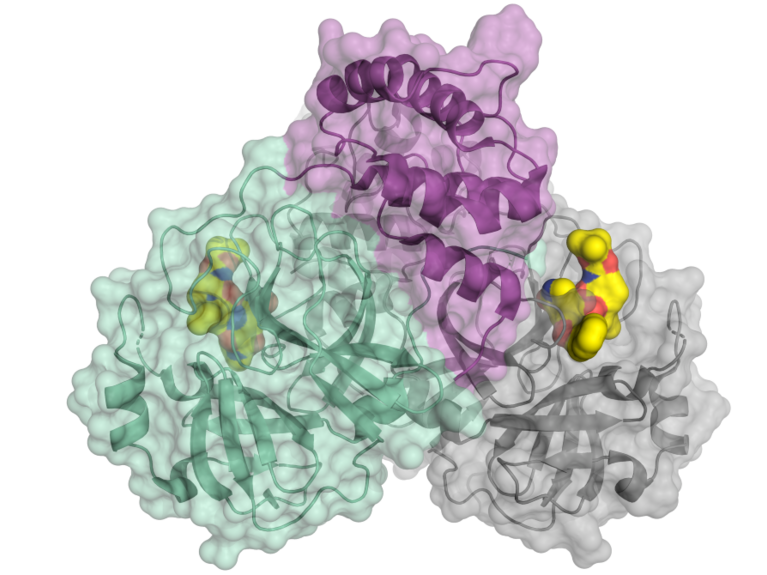
Schematic representation of the coronavirus protease. Image: HZB/H. Tabermann
Researchers working at the BESSY II X-ray source have decoded the three-dimensional structure of one of the major proteins of the novel coronavirus. Manfred Weiss from the Helmholtz-Zentrum Berlin explains how this could speed up the search for a drug to treat the disease caused by the virus.
Mr. Weiss, the spread of Sars-CoV-2 continues to accelerate around the world. Researchers are working hard to develop an active substance against the novel coronavirus. How can BESSY II support these efforts?
BESSY II is a synchrotron source at the Helmholtz-Zentrum Berlin that generates high-intensity X-ray light for research purposes. Our cutting-edge instruments help decode the 3D structure of proteins. For over 15 years, we have been working with beam tubes that make it possible to test hundreds of active substance fragments in protein crystals. These form the basis so an active substance can then be developed more rapidly. Due to our special expertise, the experiment stations at BESSY II are in very high demand. Some 100 research teams regularly conduct experiments at our facility – such as the group led by Professor Rolf Hilgenfeld from the University of Lübeck, which we were able to accommodate within a few days by fast-tracking their project. It took just three days from when the researchers in Lübeck called us until their test slot.
Professor Hilgenfeld and his team succeeded in decoding the three-dimensional structure of one of the key proteins of Sars-CoV-2 at BESSY. How did the researchers go about this?
In early January, after the genome of the novel coronavirus was sequenced, Rolf Hilgenfeld and his team succeeded in deriving a specific virus protein – its main protease – from the coronavirus and determining its crystal structure. This protein is a fundamental part of the life cycle of the coronavirus, because it’s involved in the replication of the virus. The researchers from Lübeck were then able to use our instruments to decode the three-dimensional structure of the main protease.
What role does the main protease of the virus play?
When a virus attacks a cell, it completely reprograms the cell, so it produces virus proteins. These proteins initially develop in the form of a very long polyprotein with numerous chains. The protease cleaves this polyprotein into individual proteins, as this is the only way the virus can replicate and infect other cells. Now we know the three-dimensional structure of the protease. This means we basically have a blueprint we can use to develop substances that inhibit the function of the protease. It’s a huge breakthrough, and it gives researchers a starting point for developing an effective treatment. In other words, this discovery can significantly speed up the search for a drug to treat patients with the novel coronavirus.
How will these results then be used to develop an active substance?
The first step will be finding existing, established drugs that fit the active center of the protease and can block the protein. These drugs offer an obvious advantage, because they have already been approved and simply need to be expanded to include another indication. This process is well-established in the field of research and was used to find active substances against HIV, for example. Research focused on the HIV protease in that case, too. Drugs that were discovered back then are still in use today.
And what about new drugs?
Developing new substances naturally requires significantly more time and may take a number of years. This is because once you’ve identified a molecule, you first need to check whether it’s toxic or would be broken down into toxins in the body. After that come numerous clinical studies, starting with animal trials, trials with healthy humans, and then experiments with volunteers who are infected with the virus. All these processes are necessary so researchers can develop a safe medication.
Picture: HZB/Michael Setzpfandt
Dr. Manfred Weiss heads up the Macromolecular Crystallography Group at the BESSY II synchrotron source at the Helmholtz-Zentrum Berlin. Researchers from the University of Lübeck used the BESSY II high-intensity X-ray light source to decode the three-dimensional structure of one of the key proteins of SARS-CoV-2.
What exact steps are researchers taking to unravel the protein?
We developed a method at the Helmholtz-Zentrum Berlin called fragment screening. This method involves analyzing hundreds of small molecular substances and verifying which of them attach to a protease. The proteins have a complicated surface structure. It’s difficult to find molecules that are a perfect fit and impede the function of the proteins. With our approach, we essentially test thousands of possible keys until we find the one that picks the lock and can penetrate into the protease. This is a pioneering method for developing a new medication, and this is exactly what we’re discussing together with the team from Lübeck.
Professor Hilgenfeld’s paper has just been published in Science Magazine and was already available to researchers all around the world prior to that. Is it typical to have open access that’s transparent like this?
It’s standard to give the scientific community rapid access to new results in research relating to structural biology. All of the academic researchers that conduct measurements at our facility also agree to publish their results so they’re fully accessible and to make them available to other researchers. At BESSY II, we depend on scientists using our infrastructure and analyzing their specimens at our facility. When there are extremely topical and relevant research questions – like the search for an active substance against SARS-CoV-2 – we also provide fast-track access to our beam tubes – as we did for Professor Hilgenfeld and his team.
Article published in Science Magazine
Protein crystallography at BESSY II
University of Lübeck press release (in German)
Readers comments