Self-learning algorithms
How AI is revolutionizing image analysis
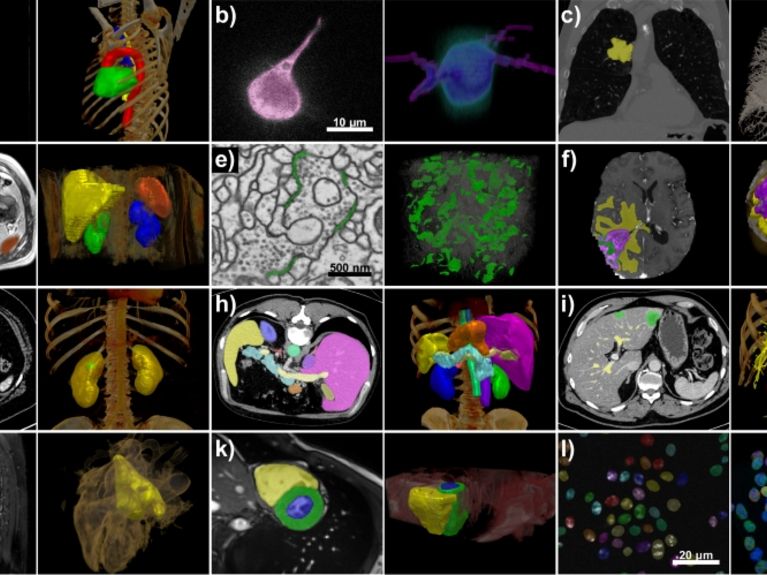
The nnU-Net program developed by Klaus Maier-Hein and his team processes a wide range of data sets and image properties. All examples are taken from the test sets of various international segmentation competitions. Image: Isensee et al. / Nature Methods
Image analysis with artificial intelligence (AI) is a major topic of the future in medicine. Medical computer scientists from Heidelberg have now developed a method with which all image data can be processed with AI - free of charge and also usable by laypersons.
Medical informatician Fabian Isensee works in the Medical Image Processing department at the German Cancer Research Center DKFZ. Image: DKFZ.
What can you see on the screen there? Is that part of a lung carcinoma? Or healthy lung tissue? A vessel? A section of a fiber of the heart muscle? Several million times a minute, the algorithm that medical computer scientist Fabian Isensee trained in Klaus Maier-Hein's team at the German Cancer Research Center (DKFZ) asks this question using a new method: What can be seen here?
For each pixel of an image from a patient's body, for example an MRI or CT image, the program searches for and finds an answer. This discrimination of which pixel belongs to which structure is called semantic segmentation in artificial intelligence (AI) research.
"In radiation therapy, doctors usually laboriously mark the cancerous tissue and surrounding structures themselves, which often takes several hours," says Fabian Isensee. "We try to relieve them of this work to some extent." To do this, they feed their program called nnU-Net with images that have already been marked. It learns from this using artificial intelligence - and applies what it has learned to new images.
The program nnU-Net
The name nnU-Net is a phonetic abbreviation for "no new net". With this, the team around Klaus Maier-Hein and Fabian Isensee wants to set an example: Their program does not require a new network or architecture; instead, it is based on a further developed standard method.
Maier-Hein and his team are not alone in this approach. The use of artificial intelligence in medical imaging is a rapidly growing field today. In many large cancer treatment centers, radiation therapists are already using algorithms to mark tumor tissue. The field is considered one of the major future topics in medicine.
Versatile benefits without time-consuming programming work
However, the new method nnU-Net developed by the experts at DKFZ has shaken up the research field in the past year - and taken a big step forward. What is special about the program is that it is not optimized for a specific segmentation task, but can be used in a variety of ways - without a lot of programming work. And that is a small revolution in the analysis of images using AI.
The first time it became clear that the Heidelberg team had initiated a paradigm shift was when they took part in an international professional competition for AI-supported image analysis, the so-called Medical Segmentation Decathlon. There, not just one data set was put up for analysis, but ten different ones: From the lungs, from the brain, from the abdominal cavity and from other body regions.
Klaus Maier-Hein heads the Medical Imaging Division at the German Cancer Research Center DKFZ. Image: DKFZ
Already this task was unusual. Because until now, it was usually the case that one had only one problem. In response to such a request, medical informaticists usually trained and refined an algorithm. "You can think of a single image type analysis task as being like a race track," explains Klaus Maier-Hein. "Typically, you have a base car - the program, how the algorithm is trained - and then with various add-ons and tuning, you specifically improve the car so that it's better in certain corners and on certain parts of the track, so it's better overall." In the decathlon, however, customized attachments are no longer an option, and so most teams simply participated with a rigid base algorithm that, on average, reasonably fit the given data.
Isensee and his colleagues, however, chose a different path. "We optimized the fine-tuning of the base car by first systematizing it and then automating it." In other words, the medical computer scientists' basic car looks at the race track - and tunes itself based on it. Next racetrack - next automatic tuning, again geared to it. And so on.
"This means we're not just faster on one track, but everywhere," says Maier-Hein. Transferred to the analysis of images, this means: Isensee and Maier-Hein first looked for commonalities that apply to all images. "We found things that can be analyzed for every image; that's the pixel edge length, for example. We then systematically created rules for this," says Isensee.
"Even medical informatics laypeople can use our free tool in a straightforward manner."
The new automatic tuning was convincing: Isensee and his team won first place by far at the Medical Segmentation Decathlon. The fact that they had indeed created a new basic method became apparent in the months that followed: "Since then, most teams have relied on nnU-Net for segmentation tasks and competitions to this day," says Isensee. The program easily prevailed in various competitions against specifically optimized solutions.
The free open-source program can also be used by laypersons
Another advantage is that even medical informatics laypersons can use the tool in an uncomplicated manner and require comparatively little technical support. Without modifying the program specifically for the task and without incorporating additional expert knowledge, one already obtains results that are comparable or even better than those of algorithms that have been processed for specific use. For broad application in everyday clinical practice, this should be a huge advantage in the future.
The team around Klaus Maier-Hein makes nnU-Net available for download free of charge as an open source tool. This means that the program is ready for immediate use. But there is still room for improvement: The configuration suggested by nnU-Net can be used by experts as a starting point for further tuning. Last year, for example, nine out of ten segmentation competitions were won by algorithms based on the program - but further optimized for the task.
Possible applications in nuclear physics, space and deep-sea research
By the way, the program can be applied not only in medicine. Basically any kind of image analysis to a good quality is possible. All that is needed is a sample data set that has already been added - so that the program can learn from it. If this is the case, nnU-Net could, for example, help marine researchers in the future to analyze seafloor photos from the deep sea; in nuclear physics, images could be broken down using an electron microscope. And in space research, for example, images from the surface of the moon could be examined. In this way, the program could develop into a kind of "AI all-rounder" in research.
Readers comments